 |
|
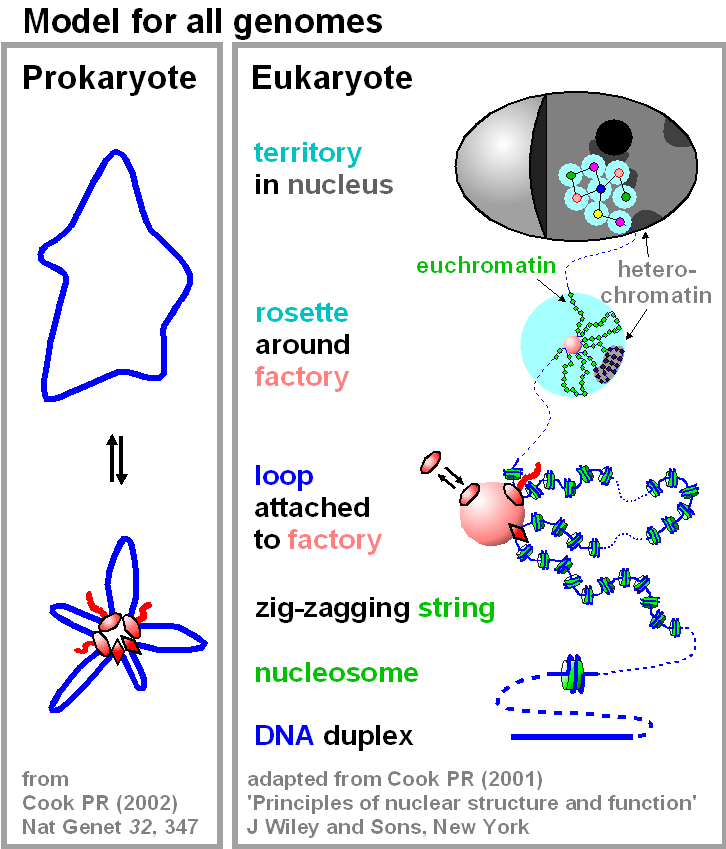
In bacteria, transcription of the circular chromosome, followed by clustering of polymerases (ovals), transcriptional activators/repressors (diamonds), and transcripts (wavy red lines), generates loops. Only one of a number of rosettes is shown, and the path of the DNA is expected to be complicated (e.g., genes a, b, c, plus p, q, and r might all be attached to rosette 1, and genes d, e, f, plus s and t to rosette 2). In eukaryotes (and specifically in a HeLa cell), DNA is coiled around the histone octamer, and runs of nucleosomes form a zig-zagging string. At the intermediate level in the hierarchy, this string is organized into loops (average contour length 86 kbp; range 5-200 kbp) by attachment to transcriptional activators/repressors (diamonds) and engaged RNA polymerases (ovals) bound to a factory. [There will be many other ties, in addition to these major ones.] 10-20 such loops (only a few are shown) form a cloud or rosette around the factory, to give a structure equivalent to that of the bacterial nucleoid. [As in the panel on the left, active transcription units that are nearest neighbours are shown bound to one factory here, but the structure is more complex; units distant on the genetic map (perhaps on different chromosomes) will sometimes bind to one factory.] In both cases, structure determines function (and vice versa); genes tethered close to a factory are more likely to initiate than distant ones. A review |
Nuclear Structure and Function Research Group |
 |
||
Images / Model for all genomes
|
|||
Top | Home | Maintained by Peter Cook | |
|||
![Transcription factories in a Hela cell [from Cook PR (1999) Science 284, 1790]](pombo.png)