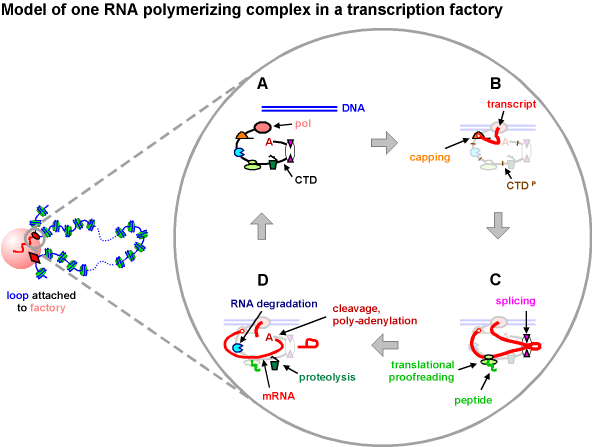
One chromatin loop attached to a transcription factory containing RNA polymerase II is shown on the left. In a HeLa cell, a typical factory of this type contains ~8 active polymerase II complexes on the surface of a ~10 MDa protein-rich core, and a blow-up of one polymerizing complex is shown on the right. We imagine that one polymerizing complex contains all the machinery necessary to create a message, and only the parts being discussed are highlighted in panels (B)-(D). Note that it remains unclear how many different components are bound to the complex at any one time, although all are shown here to be present continuously.
A. The C-terminal domain (CTD) of the largest catalytic subunit of RNA polymerase II has the potential to associate with sites involved in capping, transcript degradation, translational proofreading (involving the translational and NMD machineries), proteolysis, splicing, and polyadenylation.
B. Transcription began as the template bound to the polymerizing complex and was reeled in as the transcript was extruded; the CTD is now hyper-phosphorylated (CTDP), and a cap has been added. The 5' end of the nascent transcript will now remain tethered to the complex. [For convenience, it is shown unattached in the low-scale view on the left.]
C. The transcript continues to be extruded through a splicing site as the ribosome/NMD machinery begins proofreading the soon-to-be-spliced message. This positioning ensures that the proofreading machinery will not read introns that may contain many termination codons.
D. Once introns are removed (lariat), the transcript is cleaved and poly-adenylated, and is ready to leave for the cytoplasm; but if errors are detected by the proofreading machinery, the faulty transcript (and faulty peptide made by the ribosome) are degraded by nucleases (and proteasomes). The polymerase is still engaged, and will terminate later.
Adapted from:
Cook, P.R. (2001) 'Principles of nuclear structure and function'. J. Wiley and Sons, New York.
Iborra F.J., Escargueil, A.E., Kwek, K,Y., Akoulitchev, A., and Cook, P.R.. (2004) J. Cell Sci. 117, 899-906. [PubMed] |
![Transcription factories in a Hela cell [from Cook PR (1999) Science 284, 1790]](pombo.png)
