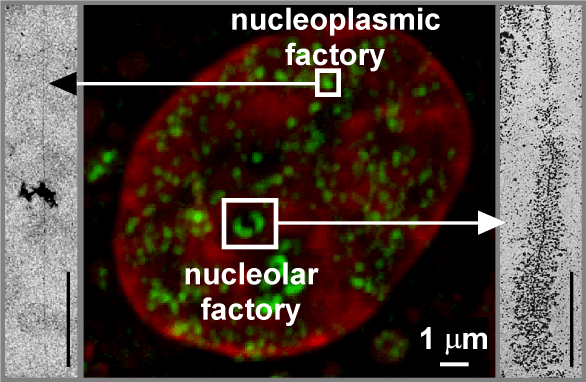
Middle. Confocal micrograph.
Cells were permeabilized, nascent RNA extended in Br-UTP, cryosectioned (100 nm), Br-RNA immunolabelled with FITC (green), nucleic acids counterstained with TOTO-3 (red), and a fluorescence image collected on a confocal microscope. Newly-made RNA (green) is concentrated in factories in the cytoplasm (made by mitochondrial polymerases), nucleoplasm (made by RNA polymerases II and III), and nucleoli (made by RNA polymerase I).
[From Cook, P.R. (1999). Science 284, 1790-1795. [PubMed] [pdf] Image kindly provided by A. Pombo.]
Sides. Electron micrographs of 'spreads' formed by tearing transcription units off underlying structures.
Left. One transcript (probably made by RNA polymerase II) is associated with DNA (which runs from top to bottom). Breaking a nucleoplasmic transcription factory like the one in the middle releases ~8 active units like this.
[From Jackson, D.A. et al., (1998). Mol. Biol.Cell 9, 1523-1536. [PubMed] [pdf]]
Right. One active (rRNA) transcription unit with ~125 transcripts; it is formed by stripping away a crescent like one of the two in the nucleolar transcription factory in the middle.
[From Miller, O.L. and Bakken, A.H. (1972). Acta Endocrinol. Suppl. 168, 155-177.] |